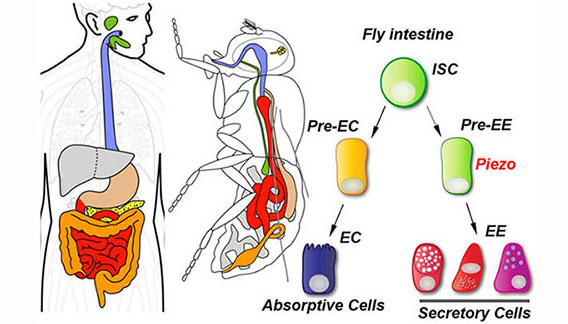
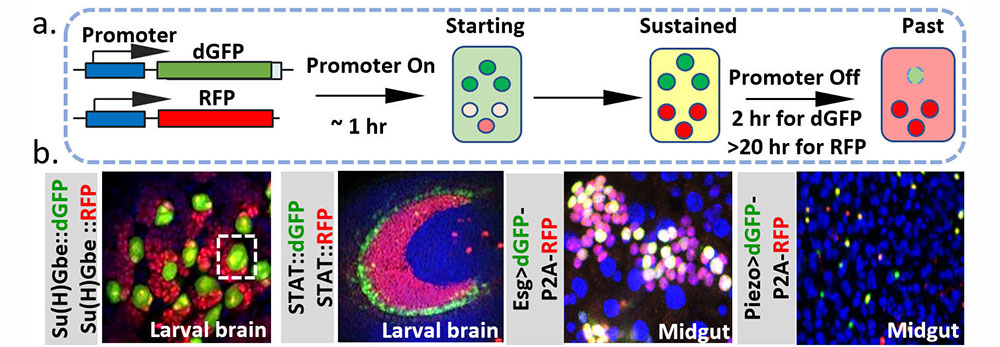
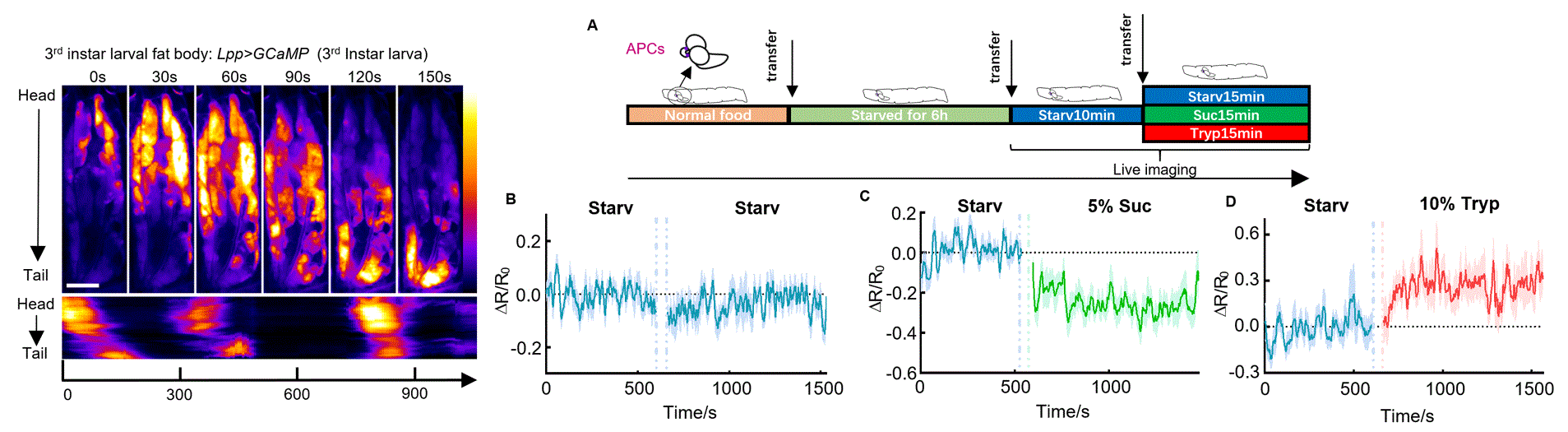
Organ Morphogenesis
As one of the three basic questions in developmental biology, studying morphogenesis not only provides the fundamental knowledge about life, but also pave the way for tissue engineering and regenerative medicine. Our lab is interested in understanding how specific cytoskeleton component is spatially and temporally regulated in individual cell to dictate the global tissue shape.
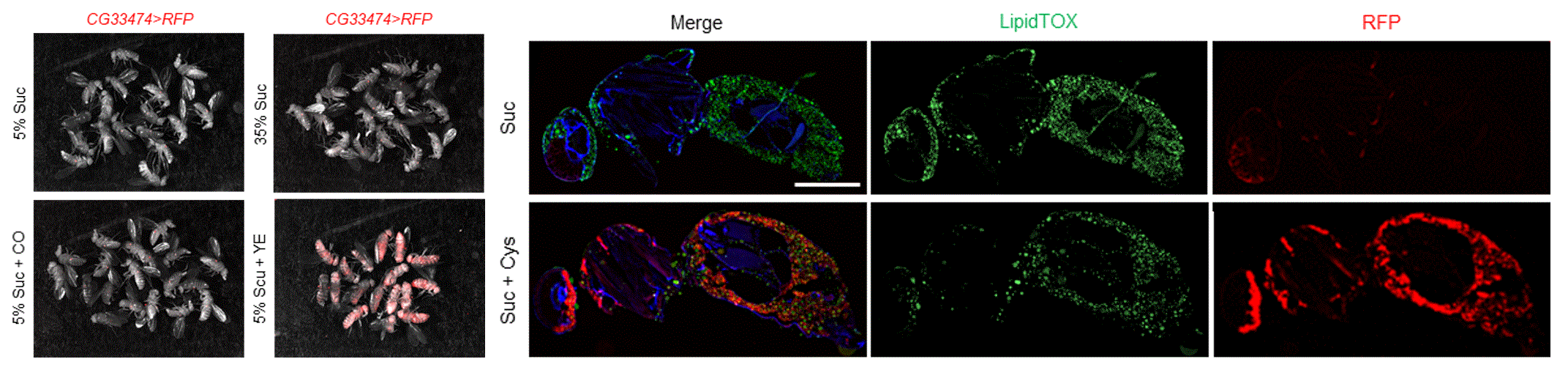
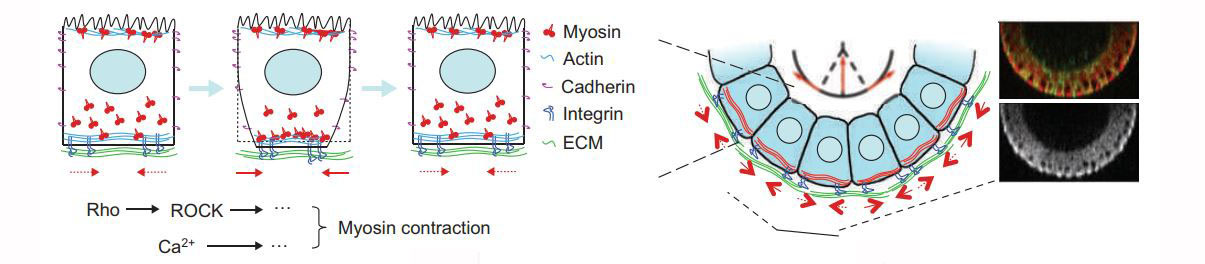
Previously, we identified a globally organized basal actomyosin contraction that promotes tissue elongation during fly oogenesis (Nat. Cell Bio. 2010).
And this global effect of actomyosin contraction can be revealed by acute removal of the basement membrane by collagenase (Mol. Bio. of the Cell, 2014).
Using this in vivo model, we are looking for new regulators that control the position and property of these contracting cells. We are also trying to identify the key switch that turn on this machinery, and test if the system can be engineered to create novel organ structures.