The true understanding of the biological system requires knowledge about exactly when, where, and how a particular molecule functions. In the past, biological studies have discovered numerous genes that regulate different biological processes. However, significant gaps still exist between these “molecular parts” and the static “phenotype” due to the lack of spatiotemporal information. Our lab uses model organisms, primarily Drosophila melanogaster, to quantitatively study the spatiotemporal dynamics of biological signals and their significance under both physiological and pathological conditions.
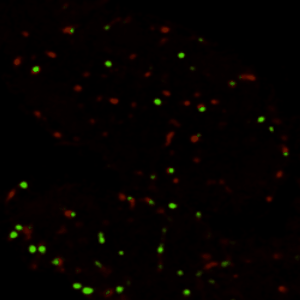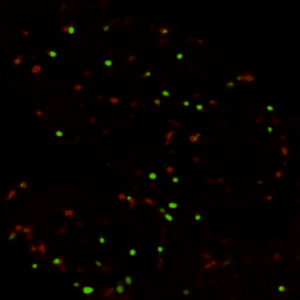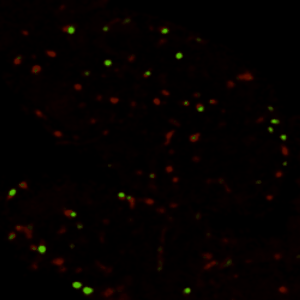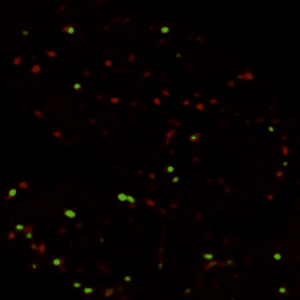
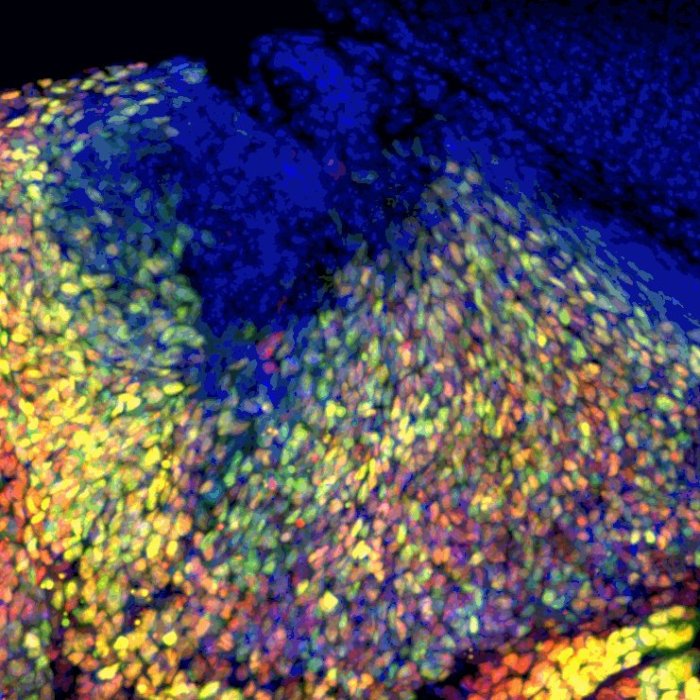
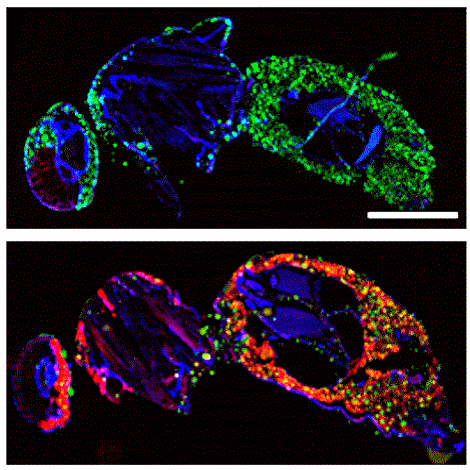
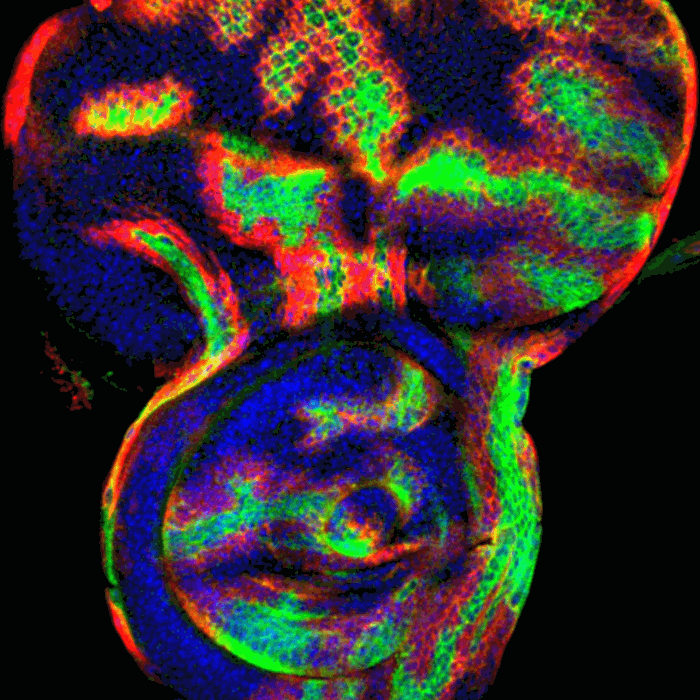
Xu Tong:Biotech Company, Meng Liu: Bengbu Medical University,Chen Zheng: Government,Sizhen Xia:Government,
.
2025
Yue Zhang; Yuchen Xia; Xinhui Wang; Yueqin Xia; Shang Wu; Jianshuang Li; Xuan Guo#; Qinghua Zhou#; Li He#. MISO regulates mitochondrial dynamics and mtDNA homeostasis by establishing membrane subdomains, Nature Cell Biology, Dec. 2025 [pdf]
Muhammad Ahmad; Shang Wu; Shengyao Luo; Wenjia Shi; Xuan Guo; Yuansheng Cao#; Norbert Perrimon#; Li He#. Dietary amino acids promote glucagon-like hormone release to generate global calcium waves in adipose tissues in Drosophila, Nature Communications, Jan. 2025 [pdf]
Chen Zheng; Jiadong Zheng; Xin Wang; Yue Zhang; Xianjue Ma#; Li He#. Two-pore-domain potassium channel Sandman regulates intestinal stem cell homeostasis and tumorigenesis in Drosophila melanogaster, Journal of Genetics and Genomics, May. 2025 [pdf]
Du Kong, Xiaoqin Li, Sihua Zhao, Chenliang Wang, Zixin Cai, Sha Song, Yifan Guo, Xiaoyu Kuang, Xianping Wang, Wenhan Liu, Peng Liu, Xiaowei Guo, Wenyan Xu, Yirong Wang, Bin Zhao, Bin Jin#, Li He#, Xianjue Ma#. Adipose tissue-secreted Spz5 promotes distal tumor progression via Toll-6-mediated Hh pathway activation in Drosophila, EMBO Journal, Jun. 2025 [pdf]
Yuxuan Sun, Boxi Sun, Xiang Cui, Weihua Li, Yue Zhang, Li He, Shutong Nong, Zhengqing Zhu, Jiyang Wu, Dongxiao Li, Xingxiang Li, Shiwu Zhang, Xiangyang Li & Mujun Li. Addressable and perceptible dynamic reprogram of ferromagnetic soft machines, Nature Communications, Mar. 2025 [pdf]
2024
Meng Liu; Li He#. Dietary cysteine and methionine promote peroxisome elevation and fat loss by induction of CG33474 expression in Drosophila adipose tissue, Cellular and Molecular Life Sciences, Dec. 2023 [pdf]
2023
He, L; Perrimon, N. Synthetic Notch receptors and their applications to study cell-cell contacts in vivo, Developmental Cell (News and Views) , Feb, 2023 [pdf]
Yuchen Xia; Yue Zhang; Yuwei Sun; Li He#. CCDC127 regulates lipid droplet homeostasis by enhancing mitochondria-ER contacts, Biochemical and Biophysical Research Communications, Nov. 2023 [pdf]
Song Song, Bomsoo Cho, Alexis T Weiner, Silas Boye Nissen, Irene Ojeda Naharros, Pablo Sanchez Bosch, Kaye Suyama, Yanhui Hu, Li He, Tanya Svinkina, Namrata D Udeshi, Steven A Carr, Norbert Perrimon & Jeffrey D Axelrod. Protein phosphatase 1 regulates core PCP signaling, EMBO reports, Dec. 2023 [pdf]
2020
Ahmad, M, He, L, Perrimon, N. Regulation of insulin and adipokinetic hormone/glucagon production in flies. Wiley Interdiscip Rev Dev Biol. 2019 [pdf]
2019
He, L*#, Binari R, Huang J, Falo-Sanjuan J, Perrimon N#. In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer. Elife. 2019 May 29 [pdf]
Hunter, G. L., He, L., Perrimon, N., Charras, G., Giniger, E., and Baum, B. A role for actomyosin contractility in Notch signaling. BMC Biology. November 2019.[pdf]
2018
He, L.*#, Si, G., Huang, J., Samuel, A., and Perrimon, N#. Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature, 07 Feb. 2018. [pdf]
Parasram, K, Bernardon, N, Hammoud, M, Chang, H, He, L, Perrimon, N, Karpowicz, P. Intestinal Stem Cells Exhibit Conditional Circadian Clock Function. Stem Cell Reports. 2018 Nov 13.[pdf]
He, L., Ahmad, M., Perrimon, N., Mechanosensitive channels and their functions in stem cell differentiation. Experimental Cell Research [pdf]
2017
He, L., Huang, J., and Perrimon, N. (2017) Development of an optimized synthetic Notch receptor as an in vivo cell-cell contact sensor. Proceedings of the National Academy of Sciences 5467-5472[pdf]
Xu, C., Luo, J., He, L., Montell, C., and Perrimon, N. (2017) Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca2+ signaling in the Drosophila midgut. eLife. [pdf]
2016
Hunter, G. L., Hadjivasiliou, Z., Bonin, H., He, L., Perrimon, N., harras, G., and Baum, B. (2016) Coordinated control of Notch/Delta signalling and cell cycle progression drives lateral inhibition-mediated tissue patterning. Development 2305-2310. [pdf]
2015
Prasad, M., Wang, X., He, L., Cai, D., and Montell, D. J. (2015) Border cell migration: a model system for live imaging and genetic analysis of collective cell movement. Drosophila Oogenesis. Methods in Molecular Biology, vol 1328.[pdf]
Chen, C.-L., Hu, Y., Udeshi, N. D., Lau, T. Y., Wirtz-Peitz, F., He, L., Ting, A. Y., Carr, S. A., and Perrimon, N. (2015) Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proceedings of the National Academy of Sciences 112.[pdf]
Gordon, W. R., Zimmerman, B., He, L., Miles, L. J., Huang, J., Tiyanont, K., McArthur, D. G., Aster, J. C., Perrimon, N., Loparo, J. J., and Blacklow, S. C. (2015) Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Developmental Cell.[pdf]
2014
Koride, S., He, L., Xiong, L.-P., Lan, G., Montell, D. J., and Sun, S. X. (2014) Mechanochemical regulation of oscillatory follicle cell dynamics in the developing Drosophila egg chamber. Molecular Biology of the Cell.[pdf]
Cai, D., Chen, S.-C., Prasad, M., He, L., Wang, X., Choesmel-Cadamuro, V., Sawyer, J. K., Danuser, G., and Montell, D. J. (2014) Mechanical feedback through Ecadherin promotes direction sensing during collective cell migration. Cell [pdf]
2012
He, L. and Montell, D. (2012) A cellular sense of touch. Nature Cell Biology (News and Views) 14, 902-903.[pdf]
2011
He, L., Wang, X., and Montell, D. J. (2011) Shining light on Drosophila oogenesis: live imaging of egg development. Current Opinion in Genetics & Development 612-619. [pdf]
Sawyer, J. K., Choi, W.*, Jung, K.-C.*, He, L.*, Harris, N. J., and Peifer, M. (2011) A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Molecular Biology of the Cell 22, 2491-2508. [pdf]
Wu, Y. I., Wang, X., He, L., Montell, D., and Hahn, K. M. (2011) Spatiotemporal control of small GTPases with light using the LOV domain. Methods in Enzymology 497, 393-407. [pdf]
Prasad, M., Wang, X., He, L., and Montell, D. J. (2011) Border cell migration: a model system for live imaging and genetic analysis of collective cell movement. Cell Migration. Methods in Molecular Biology (Methods and Protocols) [pdf]
2010
He, L.*, Wang, X.*, Tang, H. L., and Montell, D. J. (2010) Tissue elongation requires oscillating contractions of a basal actomyosin network. Nature Cell Biology 12, 1133-1142 [pdf]
Wang, X.*, He, L.*, Wu, Y. I., Hahn, K. M., and Montell, D. J. (2010) Light mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature Cell Biology 12, 591-597 [pdf]
For prospective lab members
Postdoctoral Scientist: We welcome candidates who are interested in the following directions: functional live-cell imaging, organ development and homeostasis, intestinal stem cells, and cell mechanics. Please send your CV and a brief statement of research interests to Dr. He (lihe19@ustc.edu.cn).
Graduate Students: We are looking for highly motivated individuals who are interested in conducting research in visualization and control of the dynamic properties of biological systems in vivo.
Undergraduate Students: Undergraduate students will have independent research projects and learn different techniques of molecular biology, fly genetics, and live-cell imaging.
Please send your CV to Dr. He (lihe19@ustc.edu.cn)
Email: lihe19@ustc.edu.cn
Phone: 086-0551-63606857
Mailing:
Li He, Ph.D.
Rm 107, Life Science Bldg. USTC
#443 Huangshan Street, Hefei, Anhui, 230027 P.R.China.
何立
中国科学技术大学西校区 生命科学楼 107
安徽省 合肥市 蜀山区 黄山路 443
邮编 230027